Research Areas
 My principal research interests are to digitally capture biodiversity
information using materials from natural history collections and to
employ these data to analyze, model, and decode patterns of
environmental, morphological, and community change. The patterns provide
insights into the classification of organisms, the evolution of
phenotype, and the ecological function of traits. Each specimen is an
object that contains many associated pieces of information. This
information includes the occurrence of the specimen in space, time, or
other parameters, its internal and external anatomy, DNA, stable isotope
and chemical signatures, associated publications, images, and the
individuals who studied or collected the organisms. Natural history
collections comprise libraries of millions of biological specimens,
ranging from preserved pollinating bees to dinosaur remnants. These
collections can shed light on environmental change and morphological
evolution over long periods, illuminate how species are lost, and how
biological knowledge has evolved. As a leader and key contributor to
significant specimen digitization and data aggregation efforts, I have
helped initiate, produce, and advance important regional and global
datasets. My research efforts are also guided by the recognition that
collections serve as invaluable hubs for research on specific taxa and
are integral to our global cultural heritage.
My principal research interests are to digitally capture biodiversity
information using materials from natural history collections and to
employ these data to analyze, model, and decode patterns of
environmental, morphological, and community change. The patterns provide
insights into the classification of organisms, the evolution of
phenotype, and the ecological function of traits. Each specimen is an
object that contains many associated pieces of information. This
information includes the occurrence of the specimen in space, time, or
other parameters, its internal and external anatomy, DNA, stable isotope
and chemical signatures, associated publications, images, and the
individuals who studied or collected the organisms. Natural history
collections comprise libraries of millions of biological specimens,
ranging from preserved pollinating bees to dinosaur remnants. These
collections can shed light on environmental change and morphological
evolution over long periods, illuminate how species are lost, and how
biological knowledge has evolved. As a leader and key contributor to
significant specimen digitization and data aggregation efforts, I have
helped initiate, produce, and advance important regional and global
datasets. My research efforts are also guided by the recognition that
collections serve as invaluable hubs for research on specific taxa and
are integral to our global cultural heritage.
Much of my research addresses fundamental questions in Hymenoptera
systematics and biodiversity data sharing. However, my research also
addresses interactions and communities of organisms and thus includes
investigations involving other insects and kingdoms, including plants,
birds, and reptiles. Currently, we are investigating trait evolution
through new computational tools, new methods for sharing biodiversity
data, and new efforts for describing insect-plant interactions and
distributions for conservation.

Biocollections
Central to my research are natural history collections - massive sets of biological specimens that are housed in museums, tissue collections, herbaria, botanical gardens, and zoos, and that have been compiled over several centuries. Together, they constitute our biodiversity libraries. These collections have broad cultural significance, and more importantly, constitute enormous primary resources that support fundamental research in the biological sciences. These libraries of millions of biological specimens, ranging from preserved pollinating bees to dinosaur remnants, shed light on the past, and illuminate ways that the climate has changed, that species are lost, and how biological knowledge has evolved. They also constitute voucher repositories for specimens used in research and identification of organisms of all varieties.
Investigating Traits and Morphology via Biodiversity Data Science and Computer Vision
A key research emphasis in my lab is to develop and apply new computational imaging and data science tools for fundamental investigations in morphology, anatomy, and phenotype. This research area is exemplified by a major NSF initiative I am leading, Extending Anthophila research through image and trait digitization (Big-Bee)1. A cornerstone of this 13-institution effort is the high-resolution imaging of more than one million specimens, representing more than 5000 bee species, in 2D and 3D modalities. These data and novel computational tools we are developing in the project will enable quantitative analyses of morphological features and will furnish new automated tools that aid identification. Recent advancements in computer vision and machine learning are transforming many fields of science, including entomology2. These emerging computational methods are already yielding striking results, including impressive capacities for sorting organisms3 and quantifying abundances.
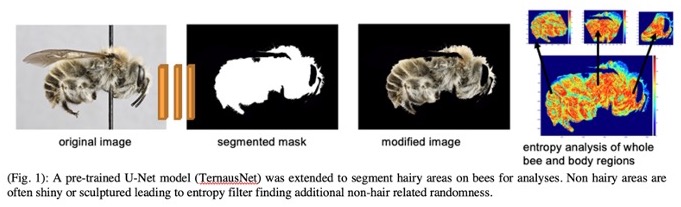
My research group is investigating multiple computer vision techniques (image segmentation, feature identification) and machine learning methods (data augmentation, transfer learning) for the quantification of bee traits, the analysis of variation between species or populations, and the development of new morphological characters for identification and systematic study. A simple but highly effective example from our current work involves the computational quantification of bee pilosity (Fig 1), a trait that may affect thermal tolerance and the ability of a species to adapt to changing climate conditions. Bee hairs have also evolved for carrying pollen, and clades differ markedly in pilosity patterns. However, hair patterns, like many morphological structures, are variable and difficult to quantify in a repeatable and scalable manner and thus exemplify the potential value that computational tools for trait analysis may contribute.
In addition to investigating new character systems, my research group is developing scalable, geographic-specific specimen classifiers using training data from images captured from natural history collections. The overarching vision of this effort is to develop methods for creating individualized models that can be used as “keys” that can be trained to encode expert domain knowledge of taxonomists or researchers.
Advancing Biodiversity Data and Knowledge Sharing
Access to biological information is dramatically reshaping the kinds
of research questions we can address and raising questions about how we
generate and manage biological data. Through my research, I examine
processes through which we capture information for research in digital
formats, use and reuse large datasets, publish and share biological
data, preserve at-risk information, and use this information in
scientific activities. My work is consequently aligned with major
research initiatives in biodiversity that seek to enable researchers to
access and exploit biodiversity data troves through global computing
systems and new technologies. Presently, I am working with a global
network of collaborators on the Extended Specimen Concept4, a
now widely accepted concept that I co-authored for the future of data
sharing. This initiative is working to promote and interconnect emerging
multimodal specimen datasets, including 3D image models5,
environmental data, and trait data (as in the Global Bee Interaction
Database6). One example of my research activity in this
overarching area is the publication “A semantically enriched taxonomic
revision of Gryonoides Dodd, 1920 (Hymenoptera, Scelionidae),
with a review of the hosts of Teleasinae.” (J. Hymenoptera Res., 2021).
This work exploits data sharing of examined material and translation
into PhenoScript, a standardized language for linking morphology with
ontology to explicitly describe characters.
Insect Distributions and Conservation
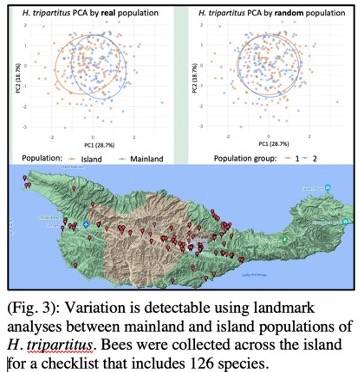
A further research focus is aimed at understanding and mitigating insect biodiversity loss. Understanding such declines requires characterizing the species distributions of insects and their host plants. My group investigates these questions by analyzing camera trap data and through biodiversity data-driven analyses of distributions. We complement these methods with basic fieldwork to enable us to evaluate hypotheses via ground-truth data. Presently, my collaborators and I are developing new techniques for analyzing co-occurrences of plants and bees from collection data, which make it possible to identify shifts or phenological mismatches. Our ongoing work includes taxonomic evaluations of bees on Santa Cruz Island, California, to produce a checklist for this scientifically valuable island ecosystem7. Species checklists are a key product of systematic research and also furnish valuable resources for informing predictions of multi-species occupancy models, ecological networks, and other community-level classifications. I am complementing these activities by developing new computer vision approaches for automating the identification and prediction of new species (Fig. 3). The methods we are developing for research can be applied to important conservation challenges, and our research engages state and federal agencies, including USFW and USGS. We have applied these methods in recent characterizations of the insects associated with the endangered California lupine plant8 and in a genome skimming analysis of herbarium specimens aimed at characterizing populations of an estuarine seablite (Madroño Journal, Accepted, 2023).
References
- Katja C. Seltmann (2021). Extending Anthophila research through image and trait digitization (Big-Bee) proposal. UC Santa Barbara: Cheadle Center for Biodiversity and Ecological Restoration. Retrieved from https://escholarship.org/uc/item/2vm761mv
- Høye, Toke T., Johanna Ärje, Kim Bjerge, Oskar LP Hansen, Alexandros Iosifidis, Florian Leese, Hjalte MR Mann, Kristian Meissner, Claus Melvad, and Jenni Raitoharju. “Deep learning and computer vision will transform entomology.” Proceedings of the National Academy of Sciences 118, no. 2 (2021): e2002545117.
- Spiesman, Brian J., Claudio Gratton, Richard G. Hatfield, William H. Hsu, Sarina Jepsen, Brian McCornack, Krushi Patel, and Guanghui Wang. “Assessing the potential for deep learning and computer vision to identify bumble bee species from images.” Scientific reports 11, no. 1 (2021): 1-10.
- James Lendemer, Barbara Thiers, Anna K Monfils, Jennifer Zaspel, Elizabeth R Ellwood, Andrew Bentley, Katherine LeVan, John Bates, David Jennings, Dori Contreras, Laura Lagomarsino, Paula Mabee, Linda S Ford, Robert Guralnick, Robert E Gropp, Marcy Revelez, Neil Cobb, Katja Seltmann, M Catherine 5. Aime, The Extended Specimen Network: A Strategy to Enhance US Biodiversity Collections, Promote Research and Education, BioScience, Volume 70, Issue 1, January 2020, Pages 23–30, https://doi.org/10.1093/biosci/biz140
- Katja C. Seltmann, Jorrit H. Poelen, UCSB Natural History Collections, & Big Bee Community. (2023). Halictus ligatus Halictidae UCSB-IZC00036803 [Data set]. Zenodo. https://doi.org/10.5281/zenodo.7510912
- Katja C. Seltmann. (2022). Global Bee Interaction Data (v2.02) [Data set]. Zenodo. https://doi.org/10.5281/zenodo.7315159
- Katja C. Seltmann, David Dewey, Lynn McLaren, & Charlie Thrift. (2022). Native and non-native bees (Anthophila) of Santa Cruz Island: An annotated checklist (0.03). Zenodo. https://doi.org/10.5281/zenodo.7216845
- C. I. Motta, J. C. Luong, K. C. Seltmann. Plant–arthropod interactions of an endangered California lupine. Ecology and Evolution. 2022 Mar;12(3):e8688.